Computation pipeline of Dataset for CNN and Mixuture NN
Contents
Machine Learning to predict location of ice recrystallizationMay - July 2022 UGA and IGE internship M1 Statistics and Data Sciences (SSD) Renan MANCEAUX Supervisor : Thomas CHAUVE Data Computing |
8.3. Computation pipeline of Dataset for CNN and Mixuture NN#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sys
sys.path.append("../../scripts/")
import utils
from tqdm.notebook import tqdm
import pickle
import xarray as xr
import xarrayaita.aita as xa
from collections import Counter
from scipy.spatial import distance_matrix
8.3.1. Loadind data#
name = 'CI09'
data = utils.load_data("../../data/for_learning_plus/"+name+".npy")
file = open('../../data/craft/'+name+'.xr', 'rb')
ds_data = pickle.load(file)
file.close()
8.3.2. Compute TJ coordinates, grains and distance using xarrayaita#
maps = ds_data.aita.dist2eachTJ()
data['idTJ'] = np.argmin(np.array(maps), axis=2).flatten()
data['grainId'] = np.array(ds_data.grainId).flatten()
imshape = np.shape(ds_data.grainId)
ntj = len(data.idTJ.unique())
M = ds_data.aita.TJ_map()
8.3.3. Filter TJ too close to border#
# TJ filter for border
tj_to_skip = []
for idtj in tqdm(np.unique(data.idTJ)):
coord = M[idtj][0:2]
if (coord[0]-4.5<0)|(coord[1]-4.5<0)|(coord[0]+4.5>imshape[1]-1)|(coord[1]+4.5>imshape[0]-1):
tj_to_skip.append(idtj)
8.3.4. Compute variables and maps#
8.3.4.1. Volume ratio anisotropy of TJ (single value)#
# anisotropy (volratio)
volratio_an_tj = np.zeros((ntj))
for i in tqdm(range(ntj)) :
if i in tj_to_skip :
volratio_an_tj[i] = np.nan
else:
coord = M[i][0:2]
volratio_an_tj[i] = np.array((data.volratio_an)).reshape(imshape)[(np.array(coord)[1]-0.5).astype(int),(np.array(coord)[0]-0.5).astype(int)]
8.3.4.2. Distance to other TJ#
# dist others TJ
Ma = ds_data.aita.TJ_map().T[0:2].T
dist = pd.DataFrame(distance_matrix(Ma,Ma))
dist2oTJ = np.zeros((ntj))
for i in tqdm(range(ntj)) :
if i in tj_to_skip :
dist2oTJ[i] = np.nan
else:
dist2oTJ[i] = np.array(dist[i].sort_values()[1:2])
8.3.4.3. Number of pixel of TJ grains (sum)#
# nb pixels neighbors grains
nb_pix_g = []
for id in tqdm(np.unique(ds_data.grainId)) :
nb_pix_g.append(int(sum(sum(ds_data.grainId == id))))
nb_pix_g = pd.Series(nb_pix_g,index=np.unique(ds_data.grainId).astype(int))
nb_pix_g_tj = np.zeros((ntj))
for i in tqdm(range(ntj)) :
if i in tj_to_skip :
nb_pix_g_tj[i] = np.nan
else:
idTJ = M[i][2:]
nb_pix_g_tj[i] = sum(np.array(nb_pix_g[np.array(idTJ)]))
8.3.4.4. Mapped values (schmid, diff_schmid, misangle and CraFT data) with a \(10 \times 10\) px window around TJ#
# schmid diff_schmid misangle work eqStrain eqStress act_py act_pr + RX_map
schmid_tj = np.zeros((ntj,10,10))
diff_schmid_tj = np.zeros((ntj,10,10))
misangle_tj = np.zeros((ntj,10,10))
work_tj = np.zeros((ntj,10,10))
eqStrain_tj = np.zeros((ntj,10,10))
eqStress_tj = np.zeros((ntj,10,10))
act_pr_tj = np.zeros((ntj,10,10))
act_py_tj = np.zeros((ntj,10,10))
RX_map = np.zeros((ntj,10,10))
for i in tqdm(range(ntj)) :
if i in tj_to_skip :
schmid_tj[i][:] = np.nan
diff_schmid_tj[i][:] = np.nan
misangle_tj[i][:] = np.nan
work_tj[i][:] = np.nan
eqStrain_tj[i][:] = np.nan
eqStress_tj[i][:] = np.nan
act_pr_tj[i][:] = np.nan
act_py_tj[i][:] = np.nan
RX_map[i][:] = np.nan
else:
coord = M[i][0:2]
y = np.array([])
x = np.array([])
for k in np.linspace(-4.5,4.5,10):
x = np.append(x,coord[0]+k).astype(int)
y = np.append(y,coord[1]+k).astype(int)
schmid_tj[i] = np.array(xr.DataArray(np.array(data.schmid).reshape(imshape))[y,x])
diff_schmid_tj[i] = np.array(xr.DataArray(np.array(data.diff_schmid).reshape(imshape))[y,x])
misangle_tj[i] = np.array(xr.DataArray(np.array(data.misangle).reshape(imshape))[y,x])
work_tj[i] = np.array(xr.DataArray(np.array(data.work).reshape(imshape))[y,x])
eqStrain_tj[i] = np.array(xr.DataArray(np.array(data.eqStrain).reshape(imshape))[y,x])
eqStress_tj[i] = np.array(xr.DataArray(np.array(data.eqStress).reshape(imshape))[y,x])
act_pr_tj[i] = np.array(xr.DataArray(np.array(data.act_pr).reshape(imshape))[y,x])
act_py_tj[i] = np.array(xr.DataArray(np.array(data.act_py).reshape(imshape))[y,x])
RX_map[i] = np.array(xr.DataArray(np.array(data.Y).reshape(imshape))[y,x])
8.3.4.5. RX value#
# RX class
RX = np.zeros((ntj))
for i in tqdm(range(ntj)):
if i in tj_to_skip :
RX[i] = np.nan
else:
m = RX_map[i]
if sum(sum(RX_map[i])) > 10: #10%
RX[i] = 1
8.3.5. Building xarray dataset#
ds = xr.Dataset()
ds['RX'] = xr.DataArray(RX,dims="nbtj")
ds['RX_map'] = xr.DataArray(RX_map,dims=["nbtj","y","x"])
ds['schmid'] = xr.DataArray(schmid_tj,dims=["nbtj","y","x"])
ds['diff_schmid'] = xr.DataArray(diff_schmid_tj,dims=["nbtj","y","x"])
ds['misangle'] = xr.DataArray(misangle_tj,dims=["nbtj","y","x"])
ds['work'] = xr.DataArray(work_tj,dims=["nbtj","y","x"])
ds['eqStrain'] = xr.DataArray(eqStrain_tj,dims=["nbtj","y","x"])
ds['eqStress'] = xr.DataArray(eqStress_tj,dims=["nbtj","y","x"])
ds['act_pr'] = xr.DataArray(act_pr_tj,dims=["nbtj","y","x"])
ds['act_py'] = xr.DataArray(act_py_tj,dims=["nbtj","y","x"])
ds['dist_to_1neigh'] = xr.DataArray(dist2oTJ,dims='nbtj')
ds['volratio_an'] = xr.DataArray(volratio_an_tj,dims='nbtj')
ds['sum_pix_g'] = xr.DataArray(nb_pix_g_tj,dims='nbtj')
ds.coords['coords'] = xr.DataArray(M.T[0:2],dims=['yx','nbtj'])
ds.attrs['Name'] = 'TJs_' + name
ds.attrs['NaN_items'] = tj_to_skip
ds
<xarray.Dataset>
Dimensions: (nbtj: 62, y: 10, x: 10, yx: 2)
Coordinates:
coords (yx, nbtj) float64 451.5 472.5 14.5 ... 528.5 532.5 534.5
Dimensions without coordinates: nbtj, y, x, yx
Data variables: (12/13)
RX (nbtj) float64 1.0 0.0 1.0 1.0 1.0 ... 0.0 0.0 1.0 0.0 nan
RX_map (nbtj, y, x) float64 0.0 0.0 0.0 0.0 0.0 ... nan nan nan nan
schmid (nbtj, y, x) float64 0.3144 0.3144 0.3144 ... nan nan nan
diff_schmid (nbtj, y, x) float64 0.1497 0.1497 0.1497 ... nan nan nan
misangle (nbtj, y, x) float64 1.311 1.311 1.311 1.311 ... nan nan nan
work (nbtj, y, x) float64 0.0008002 0.0009327 ... nan nan
... ...
eqStress (nbtj, y, x) float64 1.151 1.188 1.487 1.167 ... nan nan nan
act_pr (nbtj, y, x) float64 5.441e-06 6.949e-06 7.7e-06 ... nan nan
act_py (nbtj, y, x) float64 3.159e-10 2.266e-08 ... nan nan
dist_to_1neigh (nbtj) float64 21.38 21.38 141.0 41.23 ... 55.66 72.56 nan
volratio_an (nbtj) float64 0.9768 0.965 0.9502 ... 0.9998 0.9999 nan
sum_pix_g (nbtj) float64 9.549e+03 1.231e+04 ... 2.186e+04 nan
Attributes:
Name: TJs_CI09
NaN_items: [16, 61]8.3.5.1. Save dataset#
#ds.to_netcdf("../data/CNN/"+name+".xr")
8.3.6. Data view#
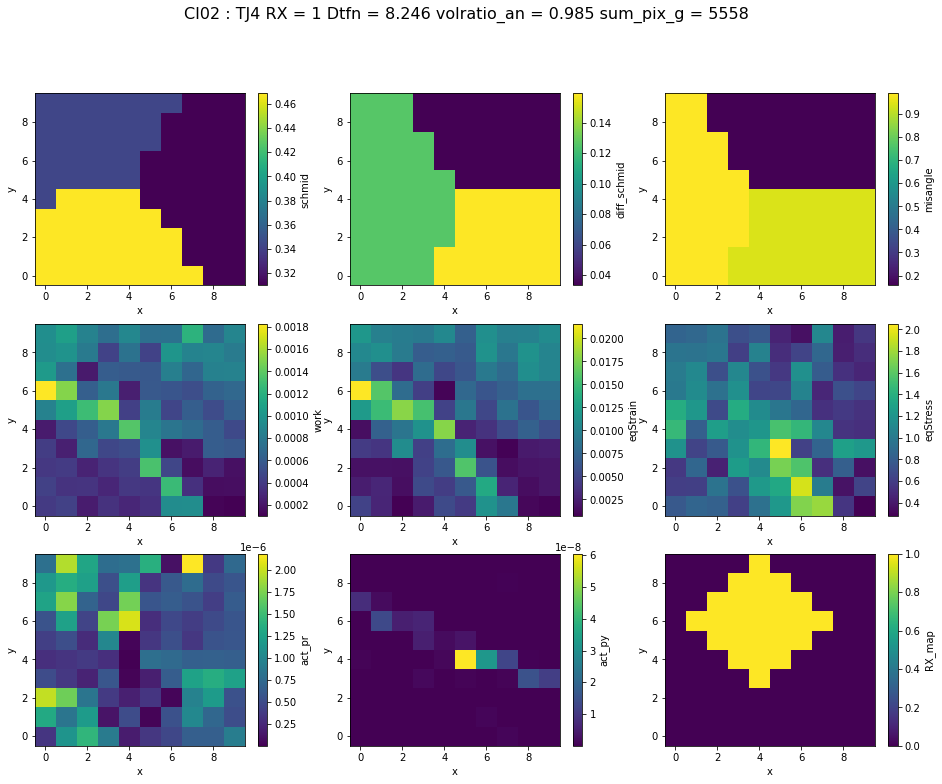
ds = xr.load_dataset("../../data/CNN/CI02.xr")
name ='CI02'
i = 4
fig = plt.figure(figsize=(16,12))
plt.subplot(331)
ds.schmid[i].plot()
plt.subplot(332)
ds.diff_schmid[i].plot()
plt.subplot(333)
ds.misangle[i].plot()
plt.subplot(334)
ds.work[i].plot()
plt.subplot(335)
ds.eqStrain[i].plot()
plt.subplot(336)
ds.eqStress[i].plot()
plt.subplot(337)
ds.act_pr[i].plot()
plt.subplot(338)
ds.act_py[i].plot()
plt.subplot(339)
ds.RX_map[i].plot()
fig.suptitle(name + " : TJ" + str(i) + " RX = "+str(int(ds.RX[i]))+" Dtfn = "+str(round(float(ds.dist_to_1neigh[i]),3))+" volratio_an = "+str(round(float(ds.volratio_an[i]),3))+" sum_pix_g = "+str(int(ds.sum_pix_g[i])),fontsize=16)
plt.show()