Apply mask#
This notebook shows how to apply a mask from a external image.
import xarrayaita.loadData_aita as lda #here are some function to build xarrayaita structure
import xarrayaita.aita as xa
import xarrayuvecs.uvecs as xu
import xarray as xr
import tifffile
import matplotlib.pyplot as plt
Load your data#
# path to data and microstructure
path_data='orientation_test.dat'
path_micro='micro_test.bmp'
data=lda.aita5col(path_data,path_micro)
data
<xarray.Dataset> Size: 100MB
Dimensions: (y: 2500, x: 1000, uvecs: 2)
Coordinates:
* x (x) float64 8kB 0.0 0.02 0.04 0.06 ... 19.92 19.94 19.96 19.98
* y (y) float64 20kB 49.98 49.96 49.94 49.92 ... 0.06 0.04 0.02 0.0
Dimensions without coordinates: uvecs
Data variables:
orientation (y, x, uvecs) float64 40MB 2.395 0.6451 5.377 ... 0.6098 0.6473
quality (y, x) int64 20MB 0 90 92 93 92 92 94 ... 96 96 96 96 97 97 96
micro (y, x) float64 20MB 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
grainId (y, x) int64 20MB 1 1 1 1 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1 1 1
Attributes:
date: b'Thursday, 19 Nov 2015, 11:24 am\r'
unit: millimeters
step_size: 0.02
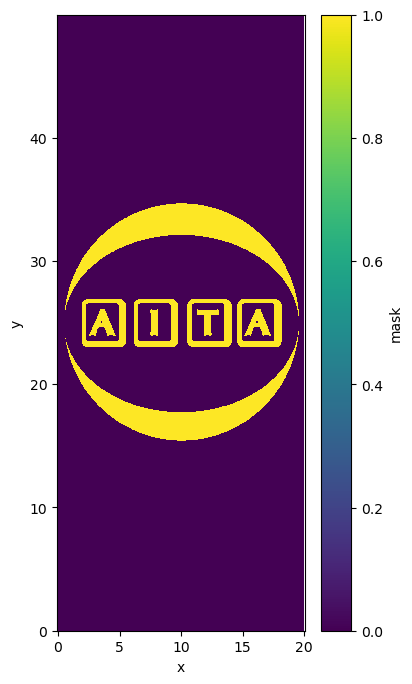
path_dat: orientation_test.datLoad mask#
This commande line load a tiff but only used the first channel. It transforms this channel into a booleen value.
data['mask']=xr.DataArray(tifffile.imread("mask_test.tiff")[...,0]>1,dims=['y','x'])
plt.figure(figsize=(4,8))
data['mask'].plot()
plt.axis('equal')
(-0.01, 19.990000000000002, -0.010000000000001563, 49.99000000000001)
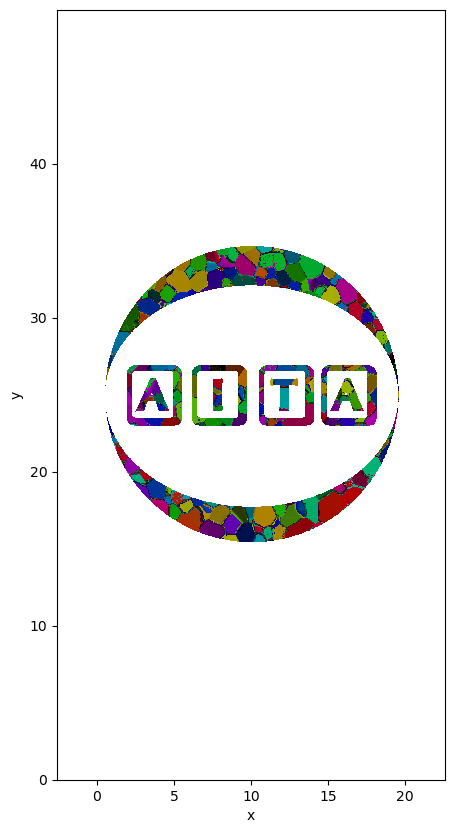
Applied the mask#
This direct applicatation of xarray power. It has nothing to do with the accessor xarrayaita.
masked_data=data.where(data.mask)
masked_data
<xarray.Dataset> Size: 120MB
Dimensions: (y: 2500, x: 1000, uvecs: 2)
Coordinates:
* x (x) float64 8kB 0.0 0.02 0.04 0.06 ... 19.92 19.94 19.96 19.98
* y (y) float64 20kB 49.98 49.96 49.94 49.92 ... 0.06 0.04 0.02 0.0
Dimensions without coordinates: uvecs
Data variables:
orientation (y, x, uvecs) float64 40MB nan nan nan nan ... nan nan nan nan
quality (y, x) float64 20MB nan nan nan nan nan ... nan nan nan nan nan
micro (y, x) float64 20MB nan nan nan nan nan ... nan nan nan nan nan
grainId (y, x) float64 20MB nan nan nan nan nan ... nan nan nan nan nan
mask (y, x) float64 20MB nan nan nan nan nan ... nan nan nan nan nan
Attributes:
date: b'Thursday, 19 Nov 2015, 11:24 am\r'
unit: millimeters
step_size: 0.02
path_dat: orientation_test.datmasked_data['FullColormap']=masked_data.orientation.uvecs.calc_colormap()
plt.figure(figsize=(5,10))
masked_data.FullColormap.plot.imshow()
plt.axis('equal')
(-0.01, 19.990000000000002, -0.010000000000001563, 49.99000000000001)